MicRhoDE: a curated database for the analysis of microbial rhodopsin diversity and evolution
Résumé
Microbial rhodopsins are a diverse group of photoactive transmembrane proteins found in all three domains of life and in viruses. Today, microbial rhodopsin research is a flourishing research field in which new understandings of rhodopsin diversity, function and evolution are contributing to broader microbiological and molecular knowledge. Here, we describe MicRhoDE, a comprehensive, high-quality and freely accessible database that facilitates analysis of the diversity and evolution of microbial rhodopsins. Rhodopsin sequences isolated from a vast array of marine and terrestrial environments were manually collected and curated. To each rhodopsin sequence are associated related metadata, including predicted spectral tuning of the protein, putative activity and function, taxonomy for sequences that can be linked to a 16S rRNA gene, sampling date and location, and supporting literature. The database currently covers 7857 aligned sequences from more than 450 environmental samples or organisms. Based on a robust phylogenetic analysis, we introduce an operational classification system with multiple phylogenetic levels ranging from superclusters to species-level operational taxonomic units. An integrated pipeline for online sequence alignment and phylogenetic tree construction is also provided. With a user-friendly interface and integrated online bioinformatics tools, this unique resource should be highly valuable for upcoming studies of the biogeography, diversity, distribution and evolution of microbial rhodopsins.
Fichier principal
 Database-2015-Boeuf-database_bav080.pdf (1.03 Mo)
Télécharger le fichier
Database-2015-Boeuf-database_bav080.pdf (1.03 Mo)
Télécharger le fichier
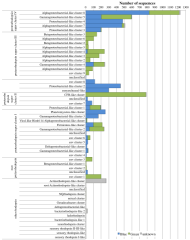 Supp. Figure 1.jpg (1.65 Mo)
Télécharger le fichier
Supplementary Figure 1.pdf (83.29 Ko)
Télécharger le fichier
Supp. Figure 1.jpg (1.65 Mo)
Télécharger le fichier
Supplementary Figure 1.pdf (83.29 Ko)
Télécharger le fichier
| Origine | Publication financée par une institution |
|---|
| Format | Figure, Image |
|---|---|
| Origine | Fichiers produits par l'(les) auteur(s) |
| Origine | Fichiers produits par l'(les) auteur(s) |
|---|

